Table of contents
In this analysis, the dataset is taken from the below link.
https://www.kaggle.com/datasets/imdevskp/corona-virus-report
Introduction
This dataset is available with Indian cases only, so the data analysis will show the different states of India and their covid cases.
In this analysis, I will show you how to fetch data from the drive and get the path of your file to perform analysis over it using the pandas Dataframe.
In this analysis, I will be visualizing the data and plotting them for better understanding.
Analysis
# importing neccesary modules
>>> import os
>>> import numpy as np
>>> import pandas as pd
>>> import matplotlib.pyplot as plt
>>> import seaborn as sns
# my data is in G-drive, to access it, we need to mount our drive
>>> from google.colab import drive
>>> drive.mount('/content/drive')
# path of my .csv file
>>> dirname='drive/My Drive/archive/complete.csv'
>>> pip install geopandas
# geopandas library makes us enable to work with geospatial data
>>> import geopandas as gpd
>>> from shapely.geometry import Point, polygon
>>> import descartes
>>> df = pd.read_csv(dirname)
Output is shown below:-
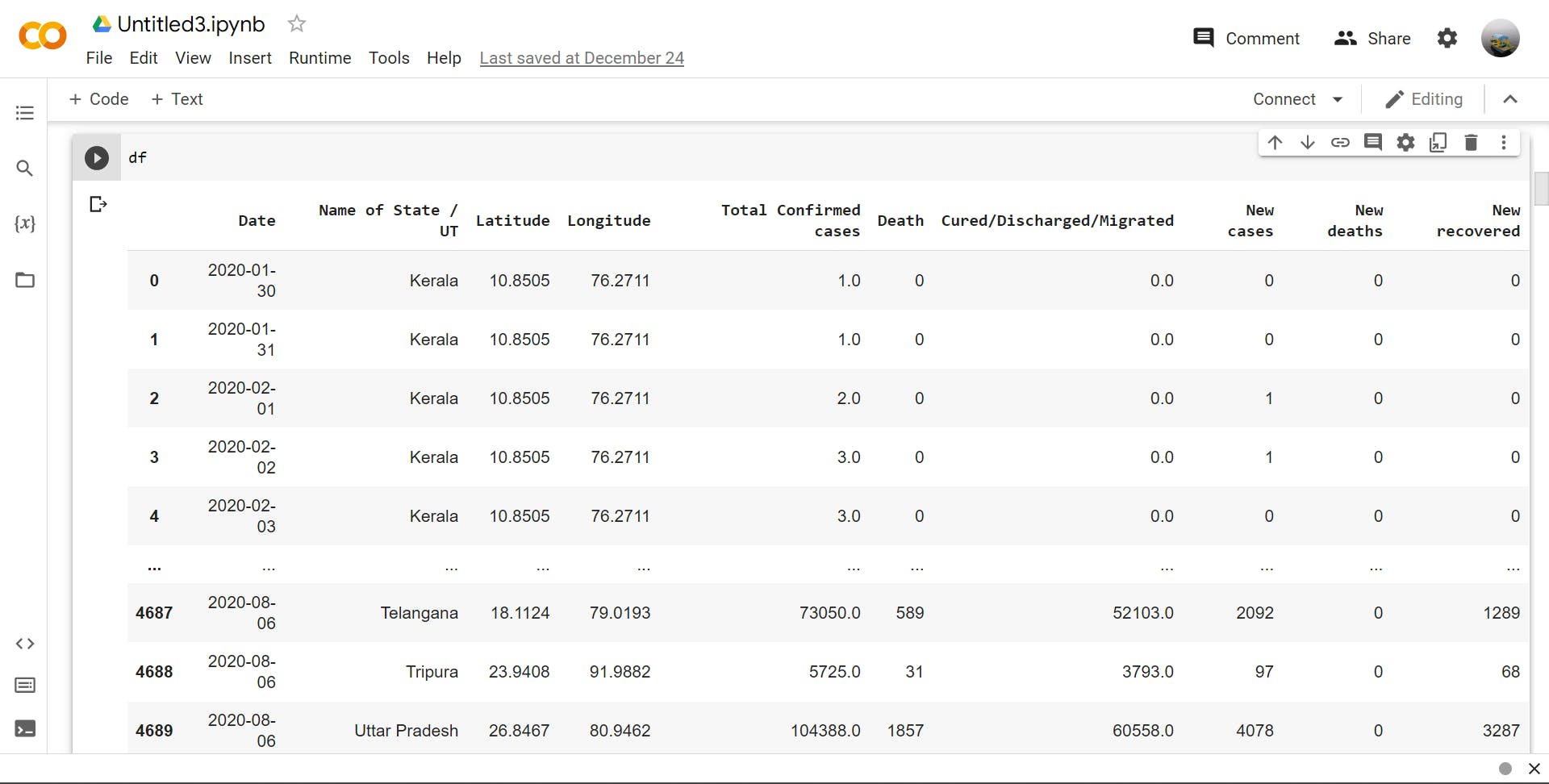
>>> df.columns
Output :-
df.columns
Index(['Date', 'Name of State / UT', 'Latitude', 'Longitude',
'Total Confirmed cases', 'Death', 'Cured/Discharged/Migrated',
'New cases', 'New deaths', 'New recovered'],
dtype='object')
# to change the names of columns
>>> df = df.rename(columns={'Name of State / UT':'State','Cured/Discharged/Migrated':'Discharged','Total Confirmed cases':'conf_cases' })
Output :-
Index(['Date', 'State', 'Latitude', 'Longitude',
'conf_cases', 'Death', 'Discharged',
'New cases', 'New deaths', 'New recovered'],
dtype='object')
>>> df.shape
Output :- (4692, 10)
>>> df.dtypes
Output :-
Date object
State object
Latitude float64
Longitude float64
conf_cases float64
Death object
Discharged float64
New cases int64
New deaths int64
New recovered int64
dtype: object
# to check total nan values in each columns
>>> df.isna().sum()
Output :-
Date 0
State 0
Latitude 0
Longitude 0
conf_cases 0
Death 0
Discharged 0
New cases 0
New deaths 0
New recovered 0
dtype: int64
>>> df.head(5).describe
#describe method is used to view a descriptive statistic
>>> df.groupby('Date')['conf_cases','Death','Discharged','New cases','New deaths'].sum().tail(10)
# this command will fetch bottom 10 rows/data of 5 columns specified in list, grouped by Date column with sum of all data present in them.
Output :-
Date conf_cases Death Discharged New cases New deaths
2020-07-28 383723.0 89 221944.0 7924 0
2020-07-29 391440.0 89 232277.0 7948 0
2020-07-30 400651.0 98 239755.0 10093 0
2020-07-31 411798.0 94 248615.0 11147 0
2020-08-01 422118.0 98 256158.0 10376 0
2020-08-02 431719.0 876 266883.0 9601 0
2020-08-03 441228.0 886 276809.0 9509 0
2020-08-04 450196.0 900 287030.0 8968 0
2020-08-05 457956.0 95 299356.0 9747 0
2020-08-06 468265.0 98 305521.0 10309 0
>>> df.groupby('State')['conf_cases','Discharged','New cases','New deaths'].max().head(10) #using head to shorten the output
# sum of all data present in specified columns, in above command will be fetched grouped by State (only 10 rows/data)
Output :-
conf_cases Discharged New cases New deaths
State
Andaman and Nicobar Islands 1027.0 326.0 99 0
Andhra Pradesh 186461.0 104354.0 10376 0
Arunachal Pradesh 1855.0 1210.0 147 0
Assam 50445.0 35892.0 2886 0
Bihar 64770.0 42414.0 3007 0
Chandigarh 1270.0 715.0 64 0
Chhattisgarh 10407.0 7871.0 512 0
Dadra and Nagar Haveli and Daman and Diu 1366.0 960.0 108 0
Delhi 140232.0 126116.0 6850 0
Goa 7423.0 5287.0 353 0
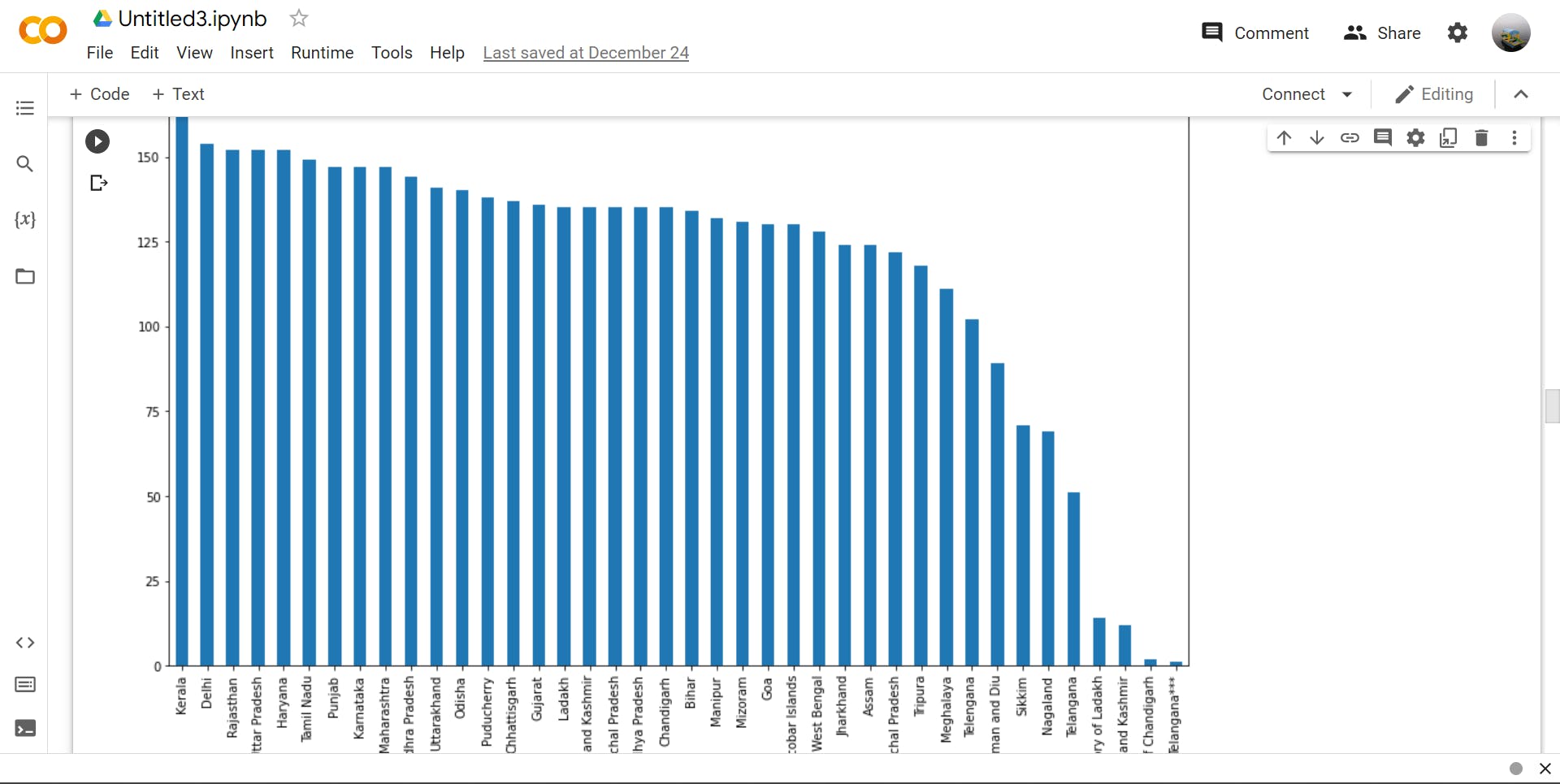
>>> df['State'].value_counts().head(5) #using head to shorten the output
#this return a Series containing counts of unique rows in the DataFrame(5 rows only)
Output :-
Kerala 186
Delhi 154
Rajasthan 152
Uttar Pradesh 152
Haryana 152
Tamil Nadu 149
>>> df['State'].value_counts().plot(kind='bar',figsize=(15,10))
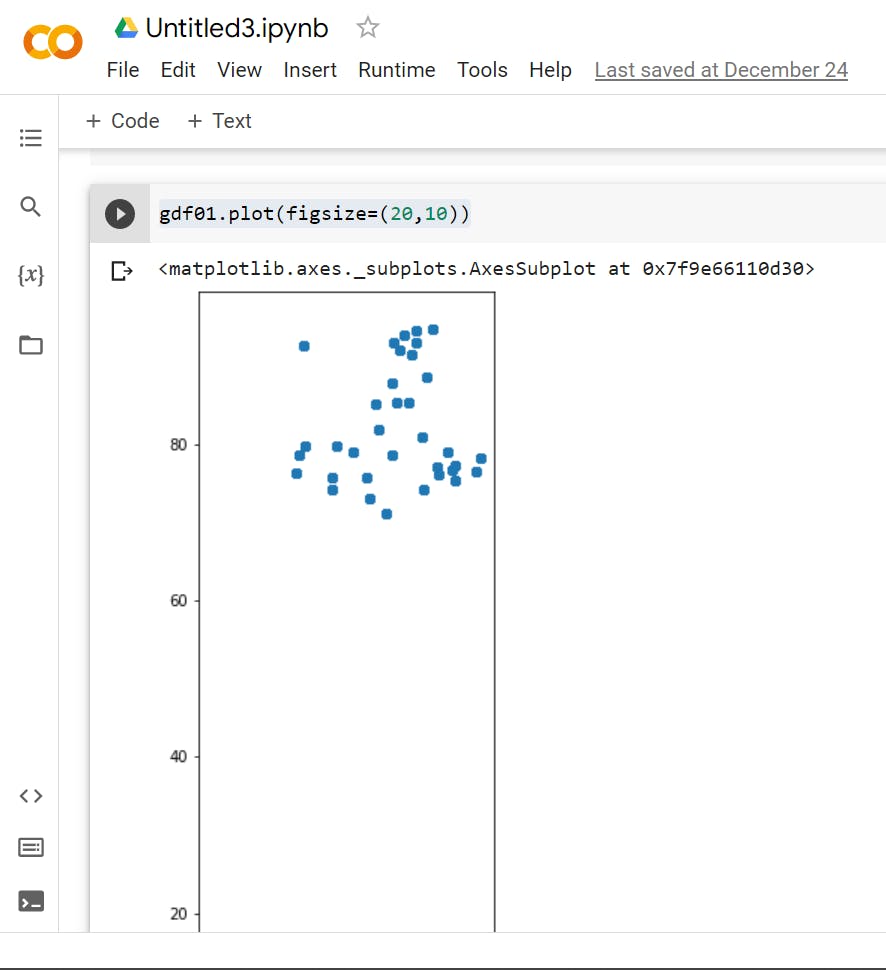
>>> gdf01 = gpd.GeoDataFrame(df,geometry=gpd.points_from_xy(df['Latitude'],df['Longitude']))
>>> gdf01.head()
# using geopandas to Generate GeometryArray of shapely Point geometries from x, y(, z) coordinates.
Ouput :-
Date State Latitude Longitude conf_cases Death Discharged New cases New deaths New recovered geometry
0 2020-01-30 Kerala 10.8505 76.2711 1.0 0 0.0 0 0 0 POINT (10.85050 76.27110)
1 2020-01-31 Kerala 10.8505 76.2711 1.0 0 0.0 0 0 0 POINT (10.85050 76.27110)
2 2020-02-01 Kerala 10.8505 76.2711 2.0 0 0.0 1 0 0 POINT (10.85050 76.27110)
3 2020-02-02 Kerala 10.8505 76.2711 3.0 0 0.0 1 0 0 POINT (10.85050 76.27110)
4 2020-02-03 Kerala 10.8505 76.2711 3.0 0 0.0 0 0 0 POINT (10.85050 76.27110)
>>> gdf01.plot(figsize=(20,10))
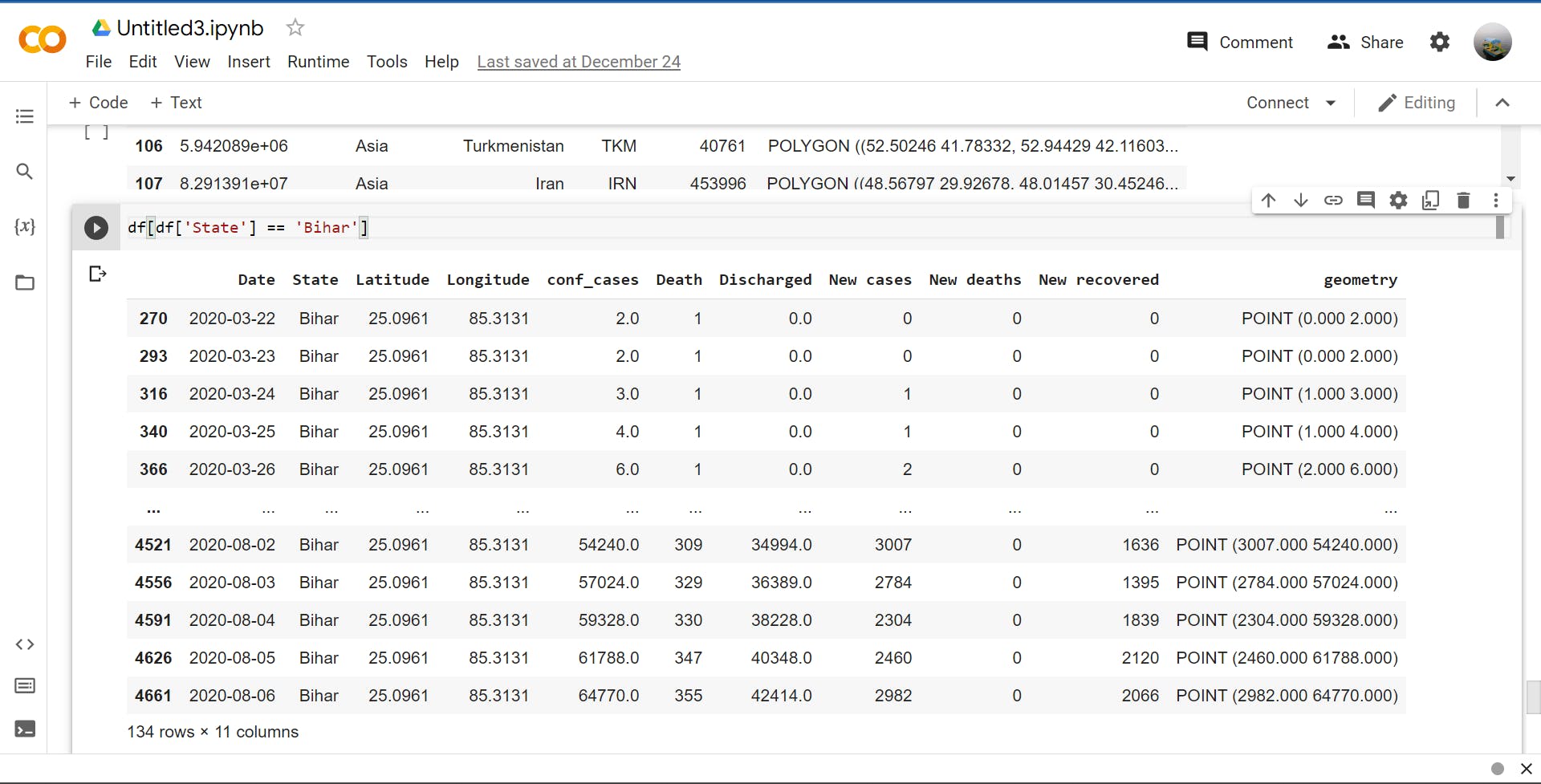
>>> df[df['State'] == 'Bihar']
# collect and show all data where state = bihar